Common Options
DataSources can be seen as "mount points" to attach actual storage to the application. Cells datasource can connect to various storage types. Each of them have specific options that are described in the following pages:
Appart from this connector type, all datasources share common options regarding how data is structured by Cells, generic Lifecycle features for data management, and some advanced settings for ETags management.
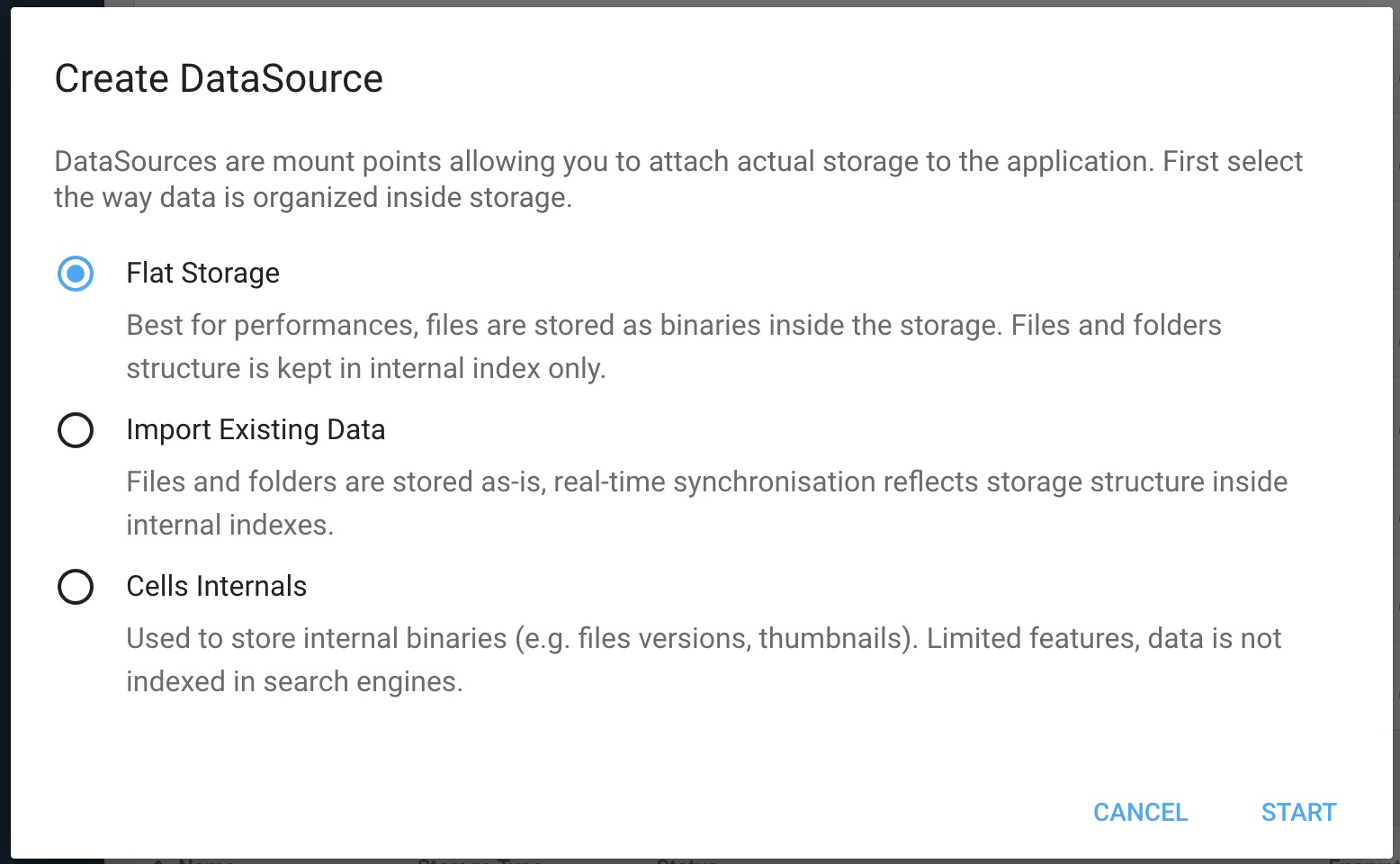
Creating a datasource
Required Options
Format
When you create a datasource, you must first select the way Cells organizes data inside the storage. Please refer to Datasource Format to have a good understanding of this feature. Quickly put, Flat Storage is better for performances, while Structured Storage is required if you wish to keep files and folders visible at the storage layer (internal datasources are specifically used for storing Thumbnails and Versions, and should not be used for your actual business data).
Identifier
Datasource identifier uniquely identifies your datasource within Cells. In the "global" tree, all files located inside this datasource will see their path start with datasource-id. Please note that this ID must be alphanumeric only, and as it is seen as an S3-BucketName internally, it must be a DNS-compliant name.
Data Lifecycle
Versioning
Setting up a versioning policy enables you to configure how Cells act with file versioning, how many versions are kept, how long we keep the old versions and many other options. You can create your own versioning policies and configure how you want to handle your versions.
File versions are stored inside the Internal DataSource named
versions
Encryption
Encryption at rest can be used to make sure files are never stored in clear format inside the storage. All files will be encrypted with a key and only be readable when decrypted with the same key.
WARNING: Keys are stored in the default database and if they ever get lost the encrypted data could not be recovered.
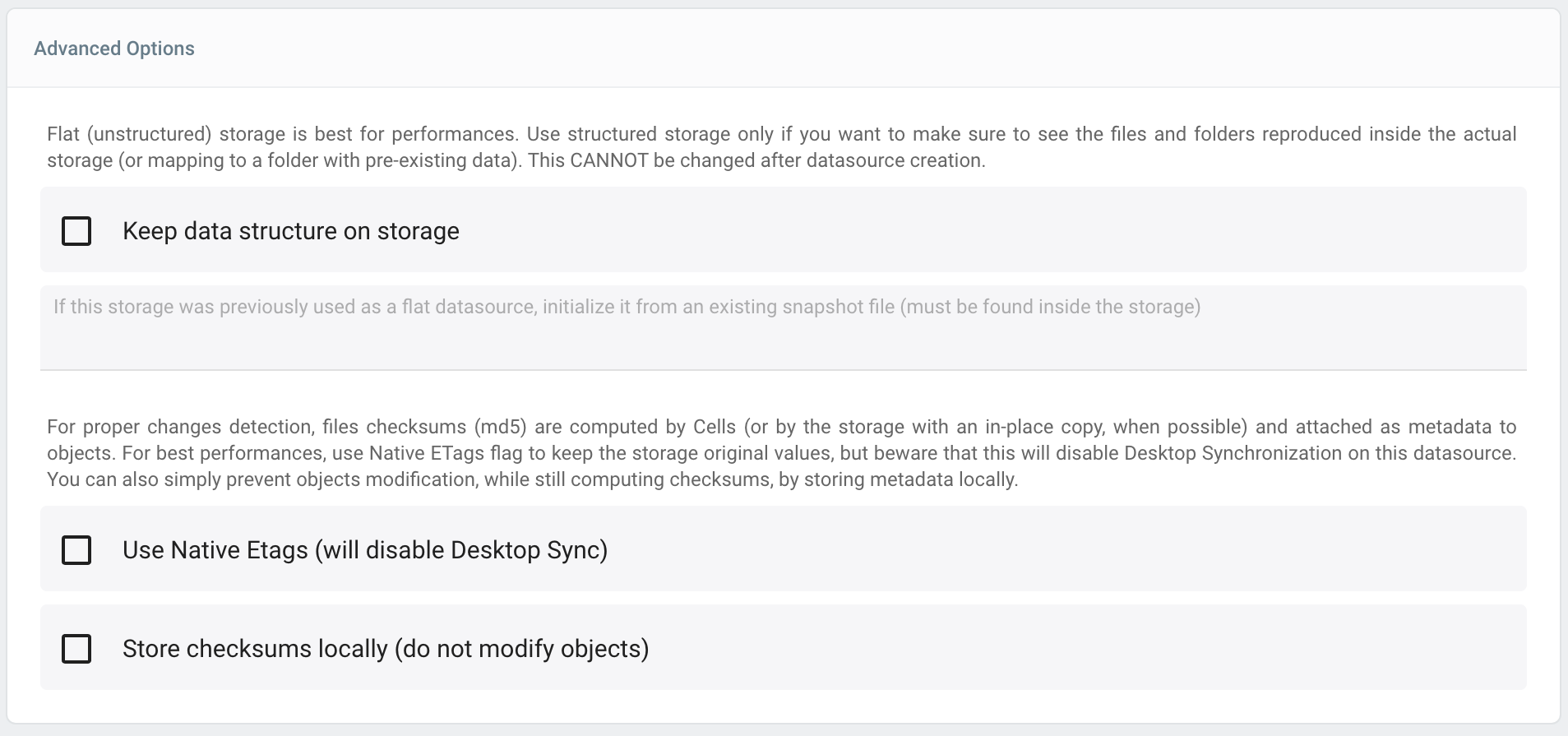
Advanced Options
Advanced options should be used for specific cases and generally touched with care!
Flat data import
When using the "Flat" storage structure, it is possible to re-import an existing datasource that has been snapshotted in a previous installation of Cells. For that, at creation, you can specify the name of the snapshot file that should be found inside the storage.
ETags Management
To achieve best performances and files reconciliation, Cells heavily relies on files checksums (ETags) that change when a file content is modified. In most cases, ETags are provided directly as MD5 hashes by the storage, but for some specific cases like files uploaded as multiparts, Cells does have to recompute them manually, and attach them as metadata to the underlying storage. This can eventually lead to bandwith consumption and may not be wanted.
Use Native ETags option will trust the storage-provided ETag value, even if it's not in the MD5 format and use it as is. As a drawback this will prevent the use of Cells Desktop Sync client as it relies on MD5 internally.
Local Checksum option will store re-computed checksums locally instead of attaching them as metadata in the storage, avoiding any modification of the original objects.